Analyzing Patient Data
We are studying inflammation in patients who have been given a new treatment for arthritis,
and need to analyze the first dozen data sets.
The data sets are stored in .csv each row holds information for a single patient, and the columns represent successive days.
The first few rows of our first file look like this:
0,0,1,3,1,2,4,7,8,3,3,3,10,5,7,4,7,7,12,18,6,13,11,11,7,7,4,6,8,8,4,4,5,7,3,4,2,3,0,0
0,1,2,1,2,1,3,2,2,6,10,11,5,9,4,4,7,16,8,6,18,4,12,5,12,7,11,5,11,3,3,5,4,4,5,5,1,1,0,1
0,1,1,3,3,2,6,2,5,9,5,7,4,5,4,15,5,11,9,10,19,14,12,17,7,12,11,7,4,2,10,5,4,2,2,3,2,2,1,1
0,0,2,0,4,2,2,1,6,7,10,7,9,13,8,8,15,10,10,7,17,4,4,7,6,15,6,4,9,11,3,5,6,3,3,4,2,3,2,1
0,1,1,3,3,1,3,5,2,4,4,7,6,5,3,10,8,10,6,17,9,14,9,7,13,9,12,6,7,7,9,6,3,2,2,4,2,0,1,1`
We want to:
- load that data into memory,
- calculate the average inflammation per day across all patients, and
- plot the result. To do all that, we'll have to learn a little bit about programming.
Objectives
- Explain what a library is, and what libraries are used for.
- Load an R library and use the things it contains.
- Read tabular data from a file into a program.
- Assign values to variables.
- Learn about data types
- Select individual values and subsections from data.
- Perform operations on arrays of data.
- Display simple graphs.
Loading Data
Words are useful, but what's more useful are the sentences and stories we use them to build. Similarly, while a lot of powerful tools are built into languages like R, even more lives in the libraries they are used to build. Importing a library is like getting a piece of lab equipment out of a storage locker and setting it up on the bench. Once it's done, we can ask the library to do things for us.
To load our inflammation data, we need to locate our data.
We will use setwd() and read.csv(). These are built-in functions in R. Let's check out the help screen.
- download the inflammation file
- put it in your working directory for these exercises
Change the current working directory to the location of the CSV file, e.g.
r
setwd("pathname")
then load the data into R
r
read.csv("data/inflammation-01.csv", header = FALSE)
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 V15 V16 V17 V18 V19 V20
## 1 0 0 1 3 1 2 4 7 8 3 3 3 10 5 7 4 7 7 12 18
## 2 0 1 2 1 2 1 3 2 2 6 10 11 5 9 4 4 7 16 8 6
## 3 0 1 1 3 3 2 6 2 5 9 5 7 4 5 4 15 5 11 9 10
## 4 0 0 2 0 4 2 2 1 6 7 10 7 9 13 8 8 15 10 10 7
## 5 0 1 1 3 3 1 3 5 2 4 4 7 6 5 3 10 8 10 6 17
## 6 0 0 1 2 2 4 2 1 6 4 7 6 6 9 9 15 4 16 18 12
## 7 0 0 2 2 4 2 2 5 5 8 6 5 11 9 4 13 5 12 10 6
## 8 0 0 1 2 3 1 2 3 5 3 7 8 8 5 10 9 15 11 18 19
## 9 0 0 0 3 1 5 6 5 5 8 2 4 11 12 10 11 9 10 17 11
## 10 0 1 1 2 1 3 5 3 5 8 6 8 12 5 13 6 13 8 16 8
## 11 0 1 0 0 4 3 3 5 5 4 5 8 7 10 13 3 7 13 15 18
## 12 0 1 0 0 3 4 2 7 8 5 2 8 11 5 5 8 14 11 6 11
## 13 0 0 2 1 4 3 6 4 6 7 9 9 3 11 6 12 4 17 13 15
## 14 0 0 0 0 1 3 1 6 6 5 5 6 3 6 13 3 10 13 9 16
## 15 0 1 2 1 1 1 4 1 5 2 3 3 10 7 13 5 7 17 6 9
## 16 0 1 1 0 1 2 4 3 6 4 7 5 5 7 5 10 7 8 18 17
## 17 0 0 0 0 2 3 6 5 7 4 3 2 10 7 9 11 12 5 12 9
## 18 0 0 0 1 2 1 4 3 6 7 4 2 12 6 12 4 14 7 8 14
## 19 0 0 2 1 2 5 4 2 7 8 4 7 11 9 8 11 15 17 11 12
## 20 0 1 2 0 1 4 3 2 2 7 3 3 12 13 11 13 6 5 9 16
## 21 0 1 1 3 1 4 4 1 8 2 2 3 12 12 10 15 13 6 5 5
## 22 0 0 2 3 2 3 2 6 3 8 7 4 6 6 9 5 12 12 8 5
## 23 0 0 0 3 4 5 1 7 7 8 2 5 12 4 10 14 5 5 17 13
## 24 0 1 1 1 1 3 3 2 6 3 9 7 8 8 4 13 7 14 11 15
## 25 0 1 1 1 2 3 5 3 6 3 7 10 3 8 12 4 12 9 15 5
## 26 0 0 2 1 3 3 2 7 4 4 3 8 12 9 12 9 5 16 8 17
## 27 0 0 1 2 4 2 2 3 5 7 10 5 5 12 3 13 4 13 7 15
## 28 0 0 1 1 1 5 1 5 2 2 4 10 4 8 14 6 15 6 12 15
## 29 0 0 2 2 3 4 6 3 7 6 4 5 8 4 7 7 6 11 12 19
## 30 0 0 0 1 4 4 6 3 8 6 4 10 12 3 3 6 8 7 17 16
## 31 0 1 1 0 3 2 4 6 8 6 2 3 11 3 14 14 12 8 8 16
## 32 0 0 2 3 3 4 5 3 6 7 10 5 10 13 14 3 8 10 9 9
## 33 0 1 2 2 2 3 6 6 6 7 6 3 11 12 13 15 15 10 14 11
## 34 0 0 2 1 3 5 6 7 5 8 9 3 12 10 12 4 12 9 13 10
## 35 0 0 1 2 4 1 5 5 2 3 4 8 8 12 5 15 9 17 7 19
## 36 0 0 0 3 1 3 6 4 3 4 8 3 4 8 3 11 5 7 10 5
## 37 0 1 2 2 2 5 5 1 4 6 3 6 5 9 6 7 4 7 16 7
## 38 0 1 1 2 3 1 5 1 2 2 5 7 6 6 5 10 6 7 17 13
## 39 0 1 0 3 2 4 1 1 5 9 10 7 12 10 9 15 12 13 13 6
## 40 0 1 1 3 1 1 5 5 3 7 2 2 3 12 4 6 8 15 16 16
## 41 0 0 0 2 2 1 3 4 5 5 6 5 5 12 13 5 7 5 11 15
## 42 0 0 1 3 3 1 2 1 8 9 2 8 10 3 8 6 10 13 11 17
## 43 0 1 1 3 4 5 2 1 3 7 9 6 10 5 8 15 11 12 15 6
## 44 0 0 1 3 1 4 3 6 7 8 5 7 11 3 6 11 6 10 6 19
## 45 0 1 1 3 3 4 4 6 3 4 9 9 7 6 8 15 12 15 6 11
## 46 0 1 2 2 4 3 1 4 8 9 5 10 10 3 4 6 7 11 16 6
## 47 0 0 2 3 4 5 4 6 2 9 7 4 9 10 8 11 16 12 15 17
## 48 0 1 1 3 1 4 6 2 8 2 10 3 11 9 13 15 5 15 6 10
## 49 0 0 1 3 2 5 1 2 7 6 6 3 12 9 4 14 4 6 12 9
## 50 0 0 1 2 3 4 5 7 5 4 10 5 12 12 5 4 7 9 18 16
## 51 0 1 2 1 1 3 5 3 6 3 10 10 11 10 13 10 13 6 6 14
## 52 0 1 2 2 3 5 2 4 5 6 8 3 5 4 3 15 15 12 16 7
## 53 0 0 0 2 4 4 5 3 3 3 10 4 4 4 14 11 15 13 10 14
## 54 0 0 2 1 1 4 4 7 2 9 4 10 12 7 6 6 11 12 9 15
## 55 0 1 2 1 1 4 5 4 4 5 9 7 10 3 13 13 8 9 17 16
## 56 0 0 1 3 2 3 6 4 5 7 2 4 11 11 3 8 8 16 5 13
## 57 0 1 1 2 2 5 1 7 4 2 5 5 4 6 6 4 16 11 14 16
## 58 0 1 1 1 4 1 6 4 6 3 6 5 6 4 14 13 13 9 12 19
## 59 0 0 0 1 4 5 6 3 8 7 9 10 8 6 5 12 15 5 10 5
## 60 0 0 1 0 3 2 5 4 8 2 9 3 3 10 12 9 14 11 13 8
## V21 V22 V23 V24 V25 V26 V27 V28 V29 V30 V31 V32 V33 V34 V35 V36 V37 V38
## 1 6 13 11 11 7 7 4 6 8 8 4 4 5 7 3 4 2 3
## 2 18 4 12 5 12 7 11 5 11 3 3 5 4 4 5 5 1 1
## 3 19 14 12 17 7 12 11 7 4 2 10 5 4 2 2 3 2 2
## 4 17 4 4 7 6 15 6 4 9 11 3 5 6 3 3 4 2 3
## 5 9 14 9 7 13 9 12 6 7 7 9 6 3 2 2 4 2 0
## 6 12 5 18 9 5 3 10 3 12 7 8 4 7 3 5 4 4 3
## 7 9 17 15 8 9 3 13 7 8 2 8 8 4 2 3 5 4 1
## 8 20 8 5 13 15 10 6 10 6 7 4 9 3 5 2 5 3 2
## 9 6 16 12 6 8 14 6 13 10 11 4 6 4 7 6 3 2 1
## 10 18 15 16 14 12 7 3 8 9 11 2 5 4 5 1 4 1 2
## 11 8 15 15 16 11 14 12 4 10 10 4 3 4 5 5 3 3 2
## 12 9 16 18 6 12 5 4 3 5 7 8 3 5 4 5 5 4 0
## 13 13 12 8 7 4 7 12 9 5 6 5 4 7 3 5 4 2 3
## 14 15 9 11 4 6 4 11 11 12 3 5 8 7 4 6 4 1 3
## 15 12 13 10 4 12 4 6 7 6 10 8 2 5 1 3 4 2 0
## 16 9 8 12 11 11 11 14 6 11 2 10 9 5 6 5 3 4 2
## 17 13 19 14 17 5 13 8 11 5 10 9 8 7 5 3 1 4 0
## 18 13 19 6 9 12 6 4 13 6 7 2 3 6 5 4 2 3 0
## 19 7 12 7 6 7 4 13 5 7 6 6 9 2 1 1 2 2 0
## 20 9 19 16 11 8 9 14 12 11 9 6 6 6 1 1 2 4 3
## 21 18 19 9 6 11 12 7 6 3 6 3 2 4 3 1 5 4 2
## 22 12 10 16 7 14 12 5 4 6 9 8 5 6 6 1 4 3 0
## 23 16 15 13 6 12 9 10 3 3 7 4 4 8 2 6 5 1 0
## 24 14 13 5 13 7 14 9 10 5 11 5 3 5 1 1 4 4 1
## 25 17 16 5 10 10 15 7 5 3 11 5 5 6 1 1 1 1 0
## 26 7 11 14 7 13 11 7 12 12 7 8 5 7 2 2 4 1 1
## 27 9 12 18 14 16 12 3 11 3 2 7 4 8 2 2 1 3 0
## 28 15 13 7 17 4 5 11 4 8 7 9 4 5 3 2 5 4 3
## 29 20 18 9 5 4 7 14 8 4 3 7 7 8 3 5 4 1 3
## 30 14 15 17 4 14 13 4 4 12 11 6 9 5 5 2 5 2 1
## 31 13 7 6 9 15 7 6 4 10 8 10 4 2 6 5 5 2 3
## 32 19 15 15 6 8 8 11 5 5 7 3 6 6 4 5 2 2 3
## 33 11 8 6 12 10 5 12 7 7 11 5 8 5 2 5 5 2 0
## 34 10 6 10 11 4 15 13 7 3 4 2 9 7 2 4 2 1 2
## 35 14 18 12 17 14 4 13 13 8 11 5 6 6 2 3 5 2 1
## 36 15 9 16 17 16 3 8 9 8 3 3 9 5 1 6 5 4 2
## 37 16 13 9 16 12 6 7 9 10 3 6 4 5 4 6 3 4 3
## 38 15 16 17 14 4 4 10 10 10 11 9 9 5 4 4 2 1 0
## 39 19 9 10 6 13 5 13 6 7 2 5 5 2 1 1 1 1 3
## 40 15 4 14 5 13 10 7 10 6 3 2 3 6 3 3 5 4 3
## 41 18 7 9 10 14 12 11 9 10 3 2 9 6 2 2 5 3 0
## 42 19 6 4 11 6 12 7 5 5 4 4 8 2 6 6 4 2 2
## 43 12 16 6 4 14 3 12 9 6 11 5 8 5 5 6 1 2 1
## 44 18 14 6 10 7 9 8 5 8 3 10 2 5 1 5 4 2 1
## 45 6 18 5 14 15 12 9 8 3 6 10 6 8 7 2 5 4 3
## 46 14 9 11 10 10 7 10 8 8 4 5 8 4 4 5 2 4 1
## 47 19 10 18 13 15 11 8 4 7 11 6 7 6 5 1 3 1 0
## 48 10 5 14 15 12 7 4 5 11 4 6 9 5 6 1 1 2 1
## 49 12 7 11 7 16 8 13 6 7 6 10 7 6 3 1 5 4 3
## 50 16 10 15 15 10 4 3 7 5 9 4 6 2 4 1 4 2 2
## 51 5 4 5 5 9 4 12 7 7 4 7 9 3 3 6 3 4 1
## 52 20 15 12 8 9 6 12 5 8 3 8 5 4 1 3 2 1 3
## 53 11 17 9 11 11 7 10 12 10 10 10 8 7 5 2 2 4 1
## 54 15 6 6 13 5 12 9 6 4 7 7 6 5 4 1 4 2 2
## 55 16 15 12 13 5 12 10 9 11 9 4 5 5 2 2 5 1 0
## 56 16 5 8 8 6 9 10 10 9 3 3 5 3 5 4 5 3 3
## 57 14 14 8 17 4 14 13 7 6 3 7 7 5 6 3 4 2 2
## 58 9 10 15 10 9 10 10 7 5 6 8 6 6 4 3 5 2 1
## 59 8 13 18 17 14 9 13 4 10 11 10 8 8 6 5 5 2 0
## 60 6 18 11 9 13 11 8 5 5 2 8 5 3 5 4 1 3 1
## V39 V40
## 1 0 0
## 2 0 1
## 3 1 1
## 4 2 1
## 5 1 1
## 6 2 1
## 7 1 1
## 8 2 1
## 9 0 0
## 10 0 0
## 11 2 1
## 12 1 1
## 13 0 1
## 14 0 0
## 15 2 0
## 16 2 0
## 17 2 1
## 18 1 0
## 19 1 0
## 20 1 1
## 21 2 0
## 22 2 0
## 23 1 0
## 24 2 0
## 25 2 1
## 26 1 0
## 27 1 1
## 28 2 1
## 29 1 0
## 30 0 1
## 31 2 1
## 32 0 0
## 33 2 1
## 34 1 1
## 35 1 1
## 36 2 0
## 37 2 1
## 38 1 0
## 39 0 1
## 40 2 1
## 41 0 1
## 42 0 0
## 43 2 0
## 44 0 1
## 45 1 1
## 46 1 0
## 47 0 0
## 48 2 1
## 49 0 0
## 50 2 1
## 51 2 0
## 52 1 0
## 53 2 1
## 54 2 1
## 55 0 1
## 56 0 1
## 57 1 1
## 58 1 1
## 59 2 0
## 60 1 0
The expression read.csv() is a function call that asks R to run the function read.csv() that belongs to base R.
read.csv() has many arguments including the name of the file we want to read, and the delimiter that separates values on a line.
When we are finished typing and press Control+Enter on Windows or Cmd + Return on Mac, the console runs our command.
Since we haven't told it to do anything else with the function's output, the console displays it.
In this case, that output is the data we just loaded.
Our call to read.csv() read the file, but didn't save the data as an object.
To do that, we need to assign the data frame to a variable.
A variable is just a name for a value, such as x, current_temperature, or subject_id.
We can create a new variable simply by assigning a value to it using <-
r
weight_kg <- 55
Once a variable has a value, we can print it:
r
weight_kg
## [1] 55
and do arithmetic with it:
r
2.2 * weight_kg
## [1] 121
We can use the function paste() to strings made up from different objects, if we separate them with commas.
r
paste("weight in pounds:", 2.2 * weight_kg)
## [1] "weight in pounds: 121"
To write lines of text to the screen, we can use cat() or writeLines()
r
string <- paste("weight in pounds:", 2.2 * weight_kg)
cat(string)
## weight in pounds: 121
r
cat(string, "\n")
## weight in pounds: 121
r
writeLines(string)
## weight in pounds: 121
In normal use however, R will print to the console an object returned by a function or operation unless we assign it to a variable. Remember, to view a variable just type the name of the variable and hit return
r
string
## [1] "weight in pounds: 121"
We can also change an object's value by assigning it a new value:
r
weight_kg <- 57.5
# weight in kilograms is now
weight_kg
## [1] 57.5
If we imagine the variable as a sticky note with a name written on it, assignment is like putting the sticky note on a particular value This means that assigning a value to one object does not change the values of other variables. For example, let's store the subject's weight in pounds in a variable
r
weight_lb <- 2.2 * weight_kg
# weight in kg...
weight_kg
## [1] 57.5
r
# ...and in pounds
weight_lb
## [1] 126.5
and then change weight_kg:
r
weight_kg <- 100
# weight in kg now...
weight_kg
## [1] 100
r
# ...and in weight pounds still
weight_lb
## [1] 126.5
Updating a Variable
Since weight_lb doesn't "remember" where its value came from, it isn't automatically updated when weight_kg changes.
This is different from the way spreadsheets work.
Challenges
Draw diagrams showing what variables refer to what values after each statement in the following program:
r
mass <- 47.5
age <- 122
mass <- mass * 2
age <- age - 20
We can also add to variable that are vectors, and update them by making them longer.
For example, if we are creating a vector of patient weights, we could update that vector using c(). c() takes any number of vectors as arguments, and combines or concatenates them, in the order supplied, into a single vector.
r
weights <- 100
weights <- c(weights, 80)
What happens here is that we take the original vector weights, and we are adding the second item to the end of the first one. We can do this over and over again to build a vector or a dataset. As we program, this may be useful to autoupdate results that we are collecting or calculating.
Now that we know how to assign things to variables, let's re-run read.csv and save its result:
r
dat <- read.csv("data/inflammation-01.csv", header = FALSE)
This statement doesn't produce any output because assignment doesn't display anything. If we want to check that our data has been loaded, we can print the variable's value:
r
dat
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 V15 V16 V17 V18 V19 V20
## 1 0 0 1 3 1 2 4 7 8 3 3 3 10 5 7 4 7 7 12 18
## 2 0 1 2 1 2 1 3 2 2 6 10 11 5 9 4 4 7 16 8 6
## 3 0 1 1 3 3 2 6 2 5 9 5 7 4 5 4 15 5 11 9 10
## 4 0 0 2 0 4 2 2 1 6 7 10 7 9 13 8 8 15 10 10 7
## 5 0 1 1 3 3 1 3 5 2 4 4 7 6 5 3 10 8 10 6 17
## 6 0 0 1 2 2 4 2 1 6 4 7 6 6 9 9 15 4 16 18 12
## 7 0 0 2 2 4 2 2 5 5 8 6 5 11 9 4 13 5 12 10 6
## 8 0 0 1 2 3 1 2 3 5 3 7 8 8 5 10 9 15 11 18 19
## 9 0 0 0 3 1 5 6 5 5 8 2 4 11 12 10 11 9 10 17 11
## 10 0 1 1 2 1 3 5 3 5 8 6 8 12 5 13 6 13 8 16 8
## 11 0 1 0 0 4 3 3 5 5 4 5 8 7 10 13 3 7 13 15 18
## 12 0 1 0 0 3 4 2 7 8 5 2 8 11 5 5 8 14 11 6 11
## 13 0 0 2 1 4 3 6 4 6 7 9 9 3 11 6 12 4 17 13 15
## 14 0 0 0 0 1 3 1 6 6 5 5 6 3 6 13 3 10 13 9 16
## 15 0 1 2 1 1 1 4 1 5 2 3 3 10 7 13 5 7 17 6 9
## 16 0 1 1 0 1 2 4 3 6 4 7 5 5 7 5 10 7 8 18 17
## 17 0 0 0 0 2 3 6 5 7 4 3 2 10 7 9 11 12 5 12 9
## 18 0 0 0 1 2 1 4 3 6 7 4 2 12 6 12 4 14 7 8 14
## 19 0 0 2 1 2 5 4 2 7 8 4 7 11 9 8 11 15 17 11 12
## 20 0 1 2 0 1 4 3 2 2 7 3 3 12 13 11 13 6 5 9 16
## 21 0 1 1 3 1 4 4 1 8 2 2 3 12 12 10 15 13 6 5 5
## 22 0 0 2 3 2 3 2 6 3 8 7 4 6 6 9 5 12 12 8 5
## 23 0 0 0 3 4 5 1 7 7 8 2 5 12 4 10 14 5 5 17 13
## 24 0 1 1 1 1 3 3 2 6 3 9 7 8 8 4 13 7 14 11 15
## 25 0 1 1 1 2 3 5 3 6 3 7 10 3 8 12 4 12 9 15 5
## 26 0 0 2 1 3 3 2 7 4 4 3 8 12 9 12 9 5 16 8 17
## 27 0 0 1 2 4 2 2 3 5 7 10 5 5 12 3 13 4 13 7 15
## 28 0 0 1 1 1 5 1 5 2 2 4 10 4 8 14 6 15 6 12 15
## 29 0 0 2 2 3 4 6 3 7 6 4 5 8 4 7 7 6 11 12 19
## 30 0 0 0 1 4 4 6 3 8 6 4 10 12 3 3 6 8 7 17 16
## 31 0 1 1 0 3 2 4 6 8 6 2 3 11 3 14 14 12 8 8 16
## 32 0 0 2 3 3 4 5 3 6 7 10 5 10 13 14 3 8 10 9 9
## 33 0 1 2 2 2 3 6 6 6 7 6 3 11 12 13 15 15 10 14 11
## 34 0 0 2 1 3 5 6 7 5 8 9 3 12 10 12 4 12 9 13 10
## 35 0 0 1 2 4 1 5 5 2 3 4 8 8 12 5 15 9 17 7 19
## 36 0 0 0 3 1 3 6 4 3 4 8 3 4 8 3 11 5 7 10 5
## 37 0 1 2 2 2 5 5 1 4 6 3 6 5 9 6 7 4 7 16 7
## 38 0 1 1 2 3 1 5 1 2 2 5 7 6 6 5 10 6 7 17 13
## 39 0 1 0 3 2 4 1 1 5 9 10 7 12 10 9 15 12 13 13 6
## 40 0 1 1 3 1 1 5 5 3 7 2 2 3 12 4 6 8 15 16 16
## 41 0 0 0 2 2 1 3 4 5 5 6 5 5 12 13 5 7 5 11 15
## 42 0 0 1 3 3 1 2 1 8 9 2 8 10 3 8 6 10 13 11 17
## 43 0 1 1 3 4 5 2 1 3 7 9 6 10 5 8 15 11 12 15 6
## 44 0 0 1 3 1 4 3 6 7 8 5 7 11 3 6 11 6 10 6 19
## 45 0 1 1 3 3 4 4 6 3 4 9 9 7 6 8 15 12 15 6 11
## 46 0 1 2 2 4 3 1 4 8 9 5 10 10 3 4 6 7 11 16 6
## 47 0 0 2 3 4 5 4 6 2 9 7 4 9 10 8 11 16 12 15 17
## 48 0 1 1 3 1 4 6 2 8 2 10 3 11 9 13 15 5 15 6 10
## 49 0 0 1 3 2 5 1 2 7 6 6 3 12 9 4 14 4 6 12 9
## 50 0 0 1 2 3 4 5 7 5 4 10 5 12 12 5 4 7 9 18 16
## 51 0 1 2 1 1 3 5 3 6 3 10 10 11 10 13 10 13 6 6 14
## 52 0 1 2 2 3 5 2 4 5 6 8 3 5 4 3 15 15 12 16 7
## 53 0 0 0 2 4 4 5 3 3 3 10 4 4 4 14 11 15 13 10 14
## 54 0 0 2 1 1 4 4 7 2 9 4 10 12 7 6 6 11 12 9 15
## 55 0 1 2 1 1 4 5 4 4 5 9 7 10 3 13 13 8 9 17 16
## 56 0 0 1 3 2 3 6 4 5 7 2 4 11 11 3 8 8 16 5 13
## 57 0 1 1 2 2 5 1 7 4 2 5 5 4 6 6 4 16 11 14 16
## 58 0 1 1 1 4 1 6 4 6 3 6 5 6 4 14 13 13 9 12 19
## 59 0 0 0 1 4 5 6 3 8 7 9 10 8 6 5 12 15 5 10 5
## 60 0 0 1 0 3 2 5 4 8 2 9 3 3 10 12 9 14 11 13 8
## V21 V22 V23 V24 V25 V26 V27 V28 V29 V30 V31 V32 V33 V34 V35 V36 V37 V38
## 1 6 13 11 11 7 7 4 6 8 8 4 4 5 7 3 4 2 3
## 2 18 4 12 5 12 7 11 5 11 3 3 5 4 4 5 5 1 1
## 3 19 14 12 17 7 12 11 7 4 2 10 5 4 2 2 3 2 2
## 4 17 4 4 7 6 15 6 4 9 11 3 5 6 3 3 4 2 3
## 5 9 14 9 7 13 9 12 6 7 7 9 6 3 2 2 4 2 0
## 6 12 5 18 9 5 3 10 3 12 7 8 4 7 3 5 4 4 3
## 7 9 17 15 8 9 3 13 7 8 2 8 8 4 2 3 5 4 1
## 8 20 8 5 13 15 10 6 10 6 7 4 9 3 5 2 5 3 2
## 9 6 16 12 6 8 14 6 13 10 11 4 6 4 7 6 3 2 1
## 10 18 15 16 14 12 7 3 8 9 11 2 5 4 5 1 4 1 2
## 11 8 15 15 16 11 14 12 4 10 10 4 3 4 5 5 3 3 2
## 12 9 16 18 6 12 5 4 3 5 7 8 3 5 4 5 5 4 0
## 13 13 12 8 7 4 7 12 9 5 6 5 4 7 3 5 4 2 3
## 14 15 9 11 4 6 4 11 11 12 3 5 8 7 4 6 4 1 3
## 15 12 13 10 4 12 4 6 7 6 10 8 2 5 1 3 4 2 0
## 16 9 8 12 11 11 11 14 6 11 2 10 9 5 6 5 3 4 2
## 17 13 19 14 17 5 13 8 11 5 10 9 8 7 5 3 1 4 0
## 18 13 19 6 9 12 6 4 13 6 7 2 3 6 5 4 2 3 0
## 19 7 12 7 6 7 4 13 5 7 6 6 9 2 1 1 2 2 0
## 20 9 19 16 11 8 9 14 12 11 9 6 6 6 1 1 2 4 3
## 21 18 19 9 6 11 12 7 6 3 6 3 2 4 3 1 5 4 2
## 22 12 10 16 7 14 12 5 4 6 9 8 5 6 6 1 4 3 0
## 23 16 15 13 6 12 9 10 3 3 7 4 4 8 2 6 5 1 0
## 24 14 13 5 13 7 14 9 10 5 11 5 3 5 1 1 4 4 1
## 25 17 16 5 10 10 15 7 5 3 11 5 5 6 1 1 1 1 0
## 26 7 11 14 7 13 11 7 12 12 7 8 5 7 2 2 4 1 1
## 27 9 12 18 14 16 12 3 11 3 2 7 4 8 2 2 1 3 0
## 28 15 13 7 17 4 5 11 4 8 7 9 4 5 3 2 5 4 3
## 29 20 18 9 5 4 7 14 8 4 3 7 7 8 3 5 4 1 3
## 30 14 15 17 4 14 13 4 4 12 11 6 9 5 5 2 5 2 1
## 31 13 7 6 9 15 7 6 4 10 8 10 4 2 6 5 5 2 3
## 32 19 15 15 6 8 8 11 5 5 7 3 6 6 4 5 2 2 3
## 33 11 8 6 12 10 5 12 7 7 11 5 8 5 2 5 5 2 0
## 34 10 6 10 11 4 15 13 7 3 4 2 9 7 2 4 2 1 2
## 35 14 18 12 17 14 4 13 13 8 11 5 6 6 2 3 5 2 1
## 36 15 9 16 17 16 3 8 9 8 3 3 9 5 1 6 5 4 2
## 37 16 13 9 16 12 6 7 9 10 3 6 4 5 4 6 3 4 3
## 38 15 16 17 14 4 4 10 10 10 11 9 9 5 4 4 2 1 0
## 39 19 9 10 6 13 5 13 6 7 2 5 5 2 1 1 1 1 3
## 40 15 4 14 5 13 10 7 10 6 3 2 3 6 3 3 5 4 3
## 41 18 7 9 10 14 12 11 9 10 3 2 9 6 2 2 5 3 0
## 42 19 6 4 11 6 12 7 5 5 4 4 8 2 6 6 4 2 2
## 43 12 16 6 4 14 3 12 9 6 11 5 8 5 5 6 1 2 1
## 44 18 14 6 10 7 9 8 5 8 3 10 2 5 1 5 4 2 1
## 45 6 18 5 14 15 12 9 8 3 6 10 6 8 7 2 5 4 3
## 46 14 9 11 10 10 7 10 8 8 4 5 8 4 4 5 2 4 1
## 47 19 10 18 13 15 11 8 4 7 11 6 7 6 5 1 3 1 0
## 48 10 5 14 15 12 7 4 5 11 4 6 9 5 6 1 1 2 1
## 49 12 7 11 7 16 8 13 6 7 6 10 7 6 3 1 5 4 3
## 50 16 10 15 15 10 4 3 7 5 9 4 6 2 4 1 4 2 2
## 51 5 4 5 5 9 4 12 7 7 4 7 9 3 3 6 3 4 1
## 52 20 15 12 8 9 6 12 5 8 3 8 5 4 1 3 2 1 3
## 53 11 17 9 11 11 7 10 12 10 10 10 8 7 5 2 2 4 1
## 54 15 6 6 13 5 12 9 6 4 7 7 6 5 4 1 4 2 2
## 55 16 15 12 13 5 12 10 9 11 9 4 5 5 2 2 5 1 0
## 56 16 5 8 8 6 9 10 10 9 3 3 5 3 5 4 5 3 3
## 57 14 14 8 17 4 14 13 7 6 3 7 7 5 6 3 4 2 2
## 58 9 10 15 10 9 10 10 7 5 6 8 6 6 4 3 5 2 1
## 59 8 13 18 17 14 9 13 4 10 11 10 8 8 6 5 5 2 0
## 60 6 18 11 9 13 11 8 5 5 2 8 5 3 5 4 1 3 1
## V39 V40
## 1 0 0
## 2 0 1
## 3 1 1
## 4 2 1
## 5 1 1
## 6 2 1
## 7 1 1
## 8 2 1
## 9 0 0
## 10 0 0
## 11 2 1
## 12 1 1
## 13 0 1
## 14 0 0
## 15 2 0
## 16 2 0
## 17 2 1
## 18 1 0
## 19 1 0
## 20 1 1
## 21 2 0
## 22 2 0
## 23 1 0
## 24 2 0
## 25 2 1
## 26 1 0
## 27 1 1
## 28 2 1
## 29 1 0
## 30 0 1
## 31 2 1
## 32 0 0
## 33 2 1
## 34 1 1
## 35 1 1
## 36 2 0
## 37 2 1
## 38 1 0
## 39 0 1
## 40 2 1
## 41 0 1
## 42 0 0
## 43 2 0
## 44 0 1
## 45 1 1
## 46 1 0
## 47 0 0
## 48 2 1
## 49 0 0
## 50 2 1
## 51 2 0
## 52 1 0
## 53 2 1
## 54 2 1
## 55 0 1
## 56 0 1
## 57 1 1
## 58 1 1
## 59 2 0
## 60 1 0
For large data sets it is convenient to use the head() to display the first few rows of data
r
head(dat)
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 V15 V16 V17 V18 V19 V20
## 1 0 0 1 3 1 2 4 7 8 3 3 3 10 5 7 4 7 7 12 18
## 2 0 1 2 1 2 1 3 2 2 6 10 11 5 9 4 4 7 16 8 6
## 3 0 1 1 3 3 2 6 2 5 9 5 7 4 5 4 15 5 11 9 10
## 4 0 0 2 0 4 2 2 1 6 7 10 7 9 13 8 8 15 10 10 7
## 5 0 1 1 3 3 1 3 5 2 4 4 7 6 5 3 10 8 10 6 17
## 6 0 0 1 2 2 4 2 1 6 4 7 6 6 9 9 15 4 16 18 12
## V21 V22 V23 V24 V25 V26 V27 V28 V29 V30 V31 V32 V33 V34 V35 V36 V37 V38
## 1 6 13 11 11 7 7 4 6 8 8 4 4 5 7 3 4 2 3
## 2 18 4 12 5 12 7 11 5 11 3 3 5 4 4 5 5 1 1
## 3 19 14 12 17 7 12 11 7 4 2 10 5 4 2 2 3 2 2
## 4 17 4 4 7 6 15 6 4 9 11 3 5 6 3 3 4 2 3
## 5 9 14 9 7 13 9 12 6 7 7 9 6 3 2 2 4 2 0
## 6 12 5 18 9 5 3 10 3 12 7 8 4 7 3 5 4 4 3
## V39 V40
## 1 0 0
## 2 0 1
## 3 1 1
## 4 2 1
## 5 1 1
## 6 2 1
BREAK * Make sure everyone has imported the data * How many rows and columns there are * What kind of data type is it?
Manipulating Data
Now that our data is in memory, we can start doing things with it.
First, let's ask what type of thing dat is:
r
class(dat)
## [1] "data.frame"
r
str(dat)
## 'data.frame': 60 obs. of 40 variables:
## $ V1 : int 0 0 0 0 0 0 0 0 0 0 ...
## $ V2 : int 0 1 1 0 1 0 0 0 0 1 ...
## $ V3 : int 1 2 1 2 1 1 2 1 0 1 ...
## $ V4 : int 3 1 3 0 3 2 2 2 3 2 ...
## $ V5 : int 1 2 3 4 3 2 4 3 1 1 ...
## $ V6 : int 2 1 2 2 1 4 2 1 5 3 ...
## $ V7 : int 4 3 6 2 3 2 2 2 6 5 ...
## $ V8 : int 7 2 2 1 5 1 5 3 5 3 ...
## $ V9 : int 8 2 5 6 2 6 5 5 5 5 ...
## $ V10: int 3 6 9 7 4 4 8 3 8 8 ...
## $ V11: int 3 10 5 10 4 7 6 7 2 6 ...
## $ V12: int 3 11 7 7 7 6 5 8 4 8 ...
## $ V13: int 10 5 4 9 6 6 11 8 11 12 ...
## $ V14: int 5 9 5 13 5 9 9 5 12 5 ...
## $ V15: int 7 4 4 8 3 9 4 10 10 13 ...
## $ V16: int 4 4 15 8 10 15 13 9 11 6 ...
## $ V17: int 7 7 5 15 8 4 5 15 9 13 ...
## $ V18: int 7 16 11 10 10 16 12 11 10 8 ...
## $ V19: int 12 8 9 10 6 18 10 18 17 16 ...
## $ V20: int 18 6 10 7 17 12 6 19 11 8 ...
## $ V21: int 6 18 19 17 9 12 9 20 6 18 ...
## $ V22: int 13 4 14 4 14 5 17 8 16 15 ...
## $ V23: int 11 12 12 4 9 18 15 5 12 16 ...
## $ V24: int 11 5 17 7 7 9 8 13 6 14 ...
## $ V25: int 7 12 7 6 13 5 9 15 8 12 ...
## $ V26: int 7 7 12 15 9 3 3 10 14 7 ...
## $ V27: int 4 11 11 6 12 10 13 6 6 3 ...
## $ V28: int 6 5 7 4 6 3 7 10 13 8 ...
## $ V29: int 8 11 4 9 7 12 8 6 10 9 ...
## $ V30: int 8 3 2 11 7 7 2 7 11 11 ...
## $ V31: int 4 3 10 3 9 8 8 4 4 2 ...
## $ V32: int 4 5 5 5 6 4 8 9 6 5 ...
## $ V33: int 5 4 4 6 3 7 4 3 4 4 ...
## $ V34: int 7 4 2 3 2 3 2 5 7 5 ...
## $ V35: int 3 5 2 3 2 5 3 2 6 1 ...
## $ V36: int 4 5 3 4 4 4 5 5 3 4 ...
## $ V37: int 2 1 2 2 2 4 4 3 2 1 ...
## $ V38: int 3 1 2 3 0 3 1 2 1 2 ...
## $ V39: int 0 0 1 2 1 2 1 2 0 0 ...
## $ V40: int 0 1 1 1 1 1 1 1 0 0 ...
The output tells us that data currently is a data frame in R. This is similar to a spreadsheet in MS Excel, that many of us are familiar with using.
data frames
The de facto data structure for most tabular data and what we use for statistics.
Data frames can have additional attributes such as rownames(), which can be useful for annotating data, like subject_id or sample_id. But most of the time they are not used.
Some additional information on data frames:
- Usually created by
read.csv()andread.table(). - Can convert to matrix with
data.matrix()(preferred) oras.matrix() - Coercion will be forced and not always what you expect.
- Can also create with
data.frame()function. - Find the number of rows and columns with
nrow(dat)andncol(dat), respectively. - Rownames are usually 1, 2, …, n.
Useful data frame functions
head()- shown first 6 rowstail()- show last 6 rowsdim()- returns the dimensionsnrow()- number of rowsncol()- number of columnsstr()- structure of each columnnames()- shows thenamesattribute for a data frame, which gives the column names.
str output tells us the dimensions and the data types (int is integer) of each column.
We can see what its shape is like this:
r
dim(dat)
## [1] 60 40
r
nrow(dat)
## [1] 60
r
ncol(dat)
## [1] 40
This tells us that data has 60 rows and 40 columns.
Indexing
If we want to get a single value from the data frame, we must provide an row and column indices for the value we want in square brackets:
r
# first value in dat
dat[1, 1]
## [1] 0
r
# middle value in dat
dat[30, 20]
## [1] 16
R indexes starting at 1. Programming languages like Fortran, MATLAB, and R start counting at 1, because that's what human beings have done for thousands of years. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that's simpler for computers to do.
An index like [30, 20] selects a single element of data frame, but we can select whole sections as well.
For example, we can select the first ten days (columns) of values for the first four patients (rows) like this:
r
dat[1:4, 1:10]
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10
## 1 0 0 1 3 1 2 4 7 8 3
## 2 0 1 2 1 2 1 3 2 2 6
## 3 0 1 1 3 3 2 6 2 5 9
## 4 0 0 2 0 4 2 2 1 6 7
The notation 1:4 means, "Start at index 1 and go to index 4."
We don't start slices at 0:
r
dat[5:10, 0:10]
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10
## 5 0 1 1 3 3 1 3 5 2 4
## 6 0 0 1 2 2 4 2 1 6 4
## 7 0 0 2 2 4 2 2 5 5 8
## 8 0 0 1 2 3 1 2 3 5 3
## 9 0 0 0 3 1 5 6 5 5 8
## 10 0 1 1 2 1 3 5 3 5 8
and we don't have to take all the values in the slice, we can use c() to select certain values or groups of values:
r
dat[c(1:10, 20:30), c(1:10, 20:30)]
## V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V20 V21 V22 V23 V24 V25 V26 V27 V28 V29
## 1 0 0 1 3 1 2 4 7 8 3 18 6 13 11 11 7 7 4 6 8
## 2 0 1 2 1 2 1 3 2 2 6 6 18 4 12 5 12 7 11 5 11
## 3 0 1 1 3 3 2 6 2 5 9 10 19 14 12 17 7 12 11 7 4
## 4 0 0 2 0 4 2 2 1 6 7 7 17 4 4 7 6 15 6 4 9
## 5 0 1 1 3 3 1 3 5 2 4 17 9 14 9 7 13 9 12 6 7
## 6 0 0 1 2 2 4 2 1 6 4 12 12 5 18 9 5 3 10 3 12
## 7 0 0 2 2 4 2 2 5 5 8 6 9 17 15 8 9 3 13 7 8
## 8 0 0 1 2 3 1 2 3 5 3 19 20 8 5 13 15 10 6 10 6
## 9 0 0 0 3 1 5 6 5 5 8 11 6 16 12 6 8 14 6 13 10
## 10 0 1 1 2 1 3 5 3 5 8 8 18 15 16 14 12 7 3 8 9
## 20 0 1 2 0 1 4 3 2 2 7 16 9 19 16 11 8 9 14 12 11
## 21 0 1 1 3 1 4 4 1 8 2 5 18 19 9 6 11 12 7 6 3
## 22 0 0 2 3 2 3 2 6 3 8 5 12 10 16 7 14 12 5 4 6
## 23 0 0 0 3 4 5 1 7 7 8 13 16 15 13 6 12 9 10 3 3
## 24 0 1 1 1 1 3 3 2 6 3 15 14 13 5 13 7 14 9 10 5
## 25 0 1 1 1 2 3 5 3 6 3 5 17 16 5 10 10 15 7 5 3
## 26 0 0 2 1 3 3 2 7 4 4 17 7 11 14 7 13 11 7 12 12
## 27 0 0 1 2 4 2 2 3 5 7 15 9 12 18 14 16 12 3 11 3
## 28 0 0 1 1 1 5 1 5 2 2 15 15 13 7 17 4 5 11 4 8
## 29 0 0 2 2 3 4 6 3 7 6 19 20 18 9 5 4 7 14 8 4
## 30 0 0 0 1 4 4 6 3 8 6 16 14 15 17 4 14 13 4 4 12
## V30
## 1 8
## 2 3
## 3 2
## 4 11
## 5 7
## 6 7
## 7 2
## 8 7
## 9 11
## 10 11
## 20 9
## 21 6
## 22 9
## 23 7
## 24 11
## 25 11
## 26 7
## 27 2
## 28 7
## 29 3
## 30 11
Here we have taken rows and columns 1 through 10 and 20 through 30.
r
dat[seq(1, 12, 3), seq(1, 20, 3)]
## V1 V4 V7 V10 V13 V16 V19
## 1 0 3 4 3 10 4 12
## 4 0 0 2 7 9 8 10
## 7 0 2 2 8 11 13 10
## 10 0 2 5 8 12 6 16
Here we have used the built-in function seq() to take regularly spaced rows and columns.
For example, we have taken rows 1, 4, 7, and 10, and columns 1, 4, 7, 10, 13, 16, and 19.
(Again, we always include the lower bound, but stop when we reach or cross the upper bound.).
Remember, 1:10 is shorthand for seq(from = 1, to = 10, by = 1).
If we want to know the average inflammation for all patients on all days, we cannot directly take the mean of a data frame. But we can take it from a matrix.
Matrix
Matrices are a special vector in R. They are not a separate type of object but simply an atomic vector with dimensions; the number of rows and columns.
r
m <- matrix(nrow = 2, ncol = 2)
m
## [,1] [,2]
## [1,] NA NA
## [2,] NA NA
r
dim(m)
## [1] 2 2
Matrices are filled column-wise.
r
m <- matrix(1:6, nrow = 2, ncol = 3)
Other ways to construct a matrix
r
m <- 1:10
dim(m) <- c(2, 5)
This takes a vector and transform into a matrix with 2 rows and 5 columns.
Another way is to bind columns or rows using cbind() and rbind().
r
x <- 1:3
y <- 10:12
cbind(x, y)
## x y
## [1,] 1 10
## [2,] 2 11
## [3,] 3 12
r
rbind(x, y)
## [,1] [,2] [,3]
## x 1 2 3
## y 10 11 12
You can also use the byrow argument to specify how the matrix is filled. From R's own documentation:
r
mdat <- matrix(c(1, 2, 3, 11, 12, 13), nrow = 2, ncol = 3, byrow = TRUE)
mdat
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 11 12 13
Lets convert our data frame to a matrix, but give it a new name:
r
mat <- data.matrix(dat)
And then take the mean of all the values:
r
mean(mat)
## [1] 6.149
There are lots of useful built-in commands that we can use in R:
r
paste("maximum inflammation:", max(mat))
## [1] "maximum inflammation: 20"
r
paste("minimum inflammation:", min(mat))
## [1] "minimum inflammation: 0"
r
paste("standard deviation:", sd(mat))
## [1] "standard deviation: 4.61479471285207"
When analyzing data, though, we often want to look at partial statistics, such as the maximum value per patient or the average value per day. One way to do this is to select the data we want to create a new temporary array, then ask it to do the calculation:
r
patient_1 <- dat[1, ] # first row, all of the columns
max(patient_1) # max inflammation for patient 1
## [1] 18
We don't actually need to store the row in a variable of its own. Instead, we can combine the selection and the method call:
r
max(dat[2, ]) # max inflammation for patient 2
## [1] 18
EXERCISES
- If
datholds our data frame of patient data, what doesdat[3:3, 4:4]produce? What aboutdat[3:3, 4:1]? Explain the results to the person sitting next to you
Functions - Operations Across Axes
What if we need the maximum inflammation for all patients, or the average for each day? As the diagram below shows, we want to perform the operation across an axis:
To support this, in R we can use the apply() function:
r
help(apply) # r ?apply
apply() allows us to repeat a function on all of the rows (MARGIN = 1), columns (2), or both(1:2) of a matrix (or higher dimensions of an array).
If each row is a patient, and we want to know each patient's average inflammation, we will need to iterate our method across all of the rows.
r
avg_inflammation <- apply(dat, 2, mean)
Some operations, such as the column-wise means have more efficient alternatives. For example rowMeans() and colMeans().
r
colMeans(dat)
## V1 V2 V3 V4 V5 V6 V7 V8 V9
## 0.0000 0.4500 1.1167 1.7500 2.4333 3.1500 3.8000 3.8833 5.2333
## V10 V11 V12 V13 V14 V15 V16 V17 V18
## 5.5167 5.9500 5.9000 8.3500 7.7333 8.3667 9.5000 9.5833 10.6333
## V19 V20 V21 V22 V23 V24 V25 V26 V27
## 11.5667 12.3500 13.2500 11.9667 11.0333 10.1667 10.0000 8.6667 9.1500
## V28 V29 V30 V31 V32 V33 V34 V35 V36
## 7.2500 7.3333 6.5833 6.0667 5.9500 5.1167 3.6000 3.3000 3.5667
## V37 V38 V39 V40
## 2.4833 1.5000 1.1333 0.5667
Challenge
- Find the maximum and minimum values for inflammation at each day (rows are patients, and columns are days).
- Save these values to a varible.
- What is the length of your new variable?
We can also create a vector of our study days (the number of columns in data)
r
tempo <- 1:40
# r
tempo <- 1:ncol(dat)
Notice that the object was named tempo instead of time. time is a R built-in function, and as good practice avoid giving existing function names to your objects.
Now that we have all this summary information, we can put it back together into a data frame that we can use for further analysis and plotting, provided they are the same length.
r
d.summary <- data.frame(tempo, avg_inflammation, min_inflammation, max_inflammation)
Plotting
The mathematician Richard Hamming once said
The purpose of computing is insight, not numbers
and the best way to develop insight is often to visualize data. Visualization deserves an entire lecture (or course) of its own, but we can explore a few features of R's base plotting package.
Lets use the average inflammation data that we saved and plot it over the study time.
r
plot(tempo, avg_inflammation)
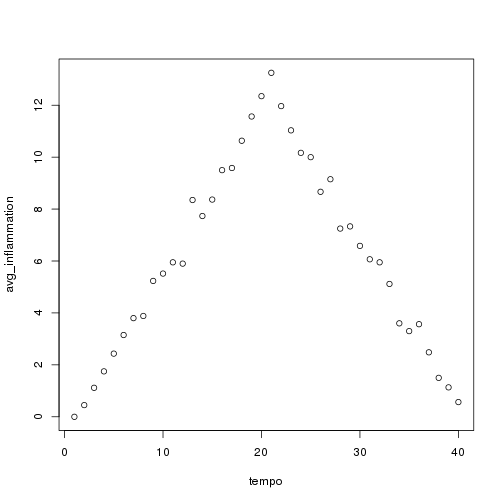
The result is roughly a linear rise and fall, which is suspicious: based on other studies, we expect a sharper rise and slower fall. Let's have a look at two other statistics:
r
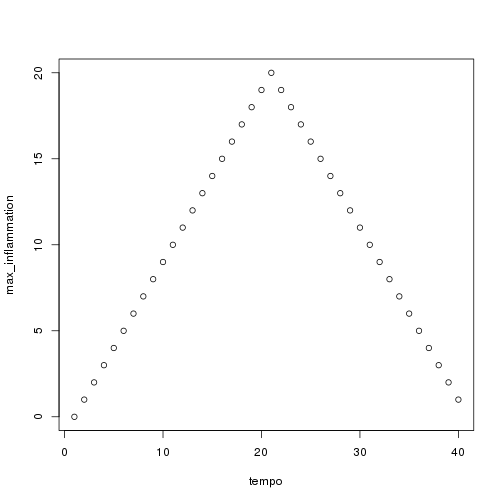
plot(tempo, max_inflammation)
r
plot(tempo, min_inflammation)
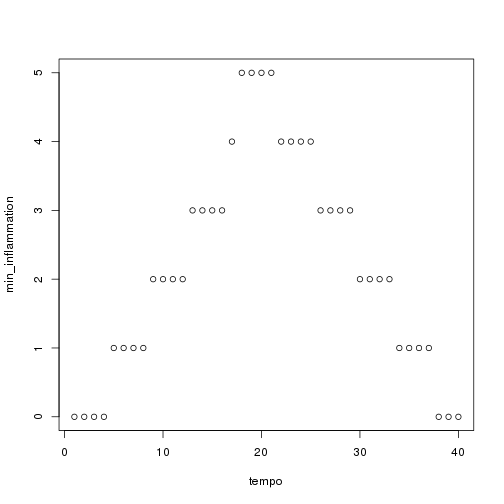
The maximum value rises and falls perfectly smoothly, while the minimum seems to be a step function. Neither result seems particularly likely, so either there's a mistake in our calculations or something is wrong with our data.
EXERCISES
- Create a plot showing the standard deviation of the inflammation data for each day across all patients.
Key Points
- Import a package into your workspace using
library("pkgname"). - The key data types in R?
- Use
variable <- valueto assign a value to a variable in order to record it in memory. - Objects are created on demand whenever a value is assigned to them.
- Use
print(something)(or justsomething) to displaysomething. - The expression
dim()gives the dimensions of a data frame or matrix. - Use
object[x, y]to select a single element from an array. - Object indices start at 1.
- Use
from:toto specify a sequence that includes the indices fromfromtoto. - Use
#to add comments to programs. - Use
mean(),max(),min()andsd()to calculate simple statistics. - Update vectors using
c() - Use base R to create simple visualizations.
Next Steps
Our work so far has convinced us that something's wrong with our first data file. We would like to check the other 11 the same way, but typing in the same commands repeatedly is tedious and error-prone. Since computers don't get bored (that we know of), we should create a way to do a complete analysis with a single command, and then figure out how to repeat that step once for each file. These operations are the subjects of the next two lessons.
